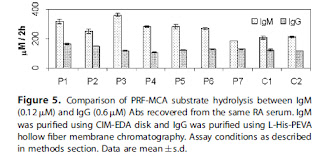
Check out the marker M (Cohn fraction IgG)
Acetate buffer, pH 5.5 Acetate buffer, pH 5.05
CIM-DEAE CIM-EDA
Again, check out marker M. Why does it look different on the right side?
Where is the BSA in CIM-EDA on the right? Did they let the gel run out? Or cut the gel off at BSA level?
So would there be as many bands under IgG in the other gels also if they let the bands resolve better?
Probably cleanest bands in lanes 1 and 2 for IgG in CIM-DEAE disk with acetate buffer, pH 5.5 (above). Would the acidic pH affect the antibody's structure/function? Can this condition be used for monoclonal IgG or IgG from sera after using the technique in J. Chrom B (2010) (Ref: post "Mixed opinions, disturbed flow) to clean up the antibodies? Is it cost-effective?
25 mM MOPS buffer, pH 6.5
CIM-DEAE
Again, check out marker M - still Cohn fraction IgG or mixture of pure IgG and BSA?
20 mM Tris buffer, pH 7.4 20 mM Tris buffer, pH 8.0
CIM-DEAE CIM-EDA
Again, check out marker M. It wasn't included in the gel for CIM-DEAE so it was cut and pasted from Fig. 2a (the acetate buffer elution gel for the same disk).
M looks even more different in the gel on the right side!
Gel image quality!!!There's still liquid between the gel and the scanner glass in the left side image!!!